The unrivalled sensitivity of Chromatrap®’s unique solid state patented technology allows for ChIP assays to be performed with as few as 1000 cells per immunoprecipitation
Usually large cell numbers are required for performing ChIP, which poses something of a challenge when dealing with limited starting material. The available techniques for tackling small samples, rare cell types or difficult cell types which provide low yield of chromatin are usually long and complex, and involve DNA amplification or the spiking of samples with chromatin from another source.
In Chromatrap®’s case, we exhibit excellent sensitivity and wide dynamic range to be able to perform ChIP from as little as 1000 cells with no difference in the level of enrichment, independent of starting cell number.
Chromatin preparation
The human endometrial carcinoma cell line Hec50 was used for chromatin extracted from 1000 cells. Cells were cultured in 96-well plates and cell counts performed on representative wells to ensure 1000 cells was achieved. Cells were then fixed in an appropriate volume of 1% formaldehyde and quenched by decanting off the formaldehyde and adding an equal volume of 0.65 M glycine. Cells were subsequently scraped in to respective amounts of ice cold PBS. Cell pellets were collected by centrifugation and cell membranes lysed in 100 µl Chromatrap® Hypotonic Buffer followed by nuclear lysis in 100 µl Chromatrap® Lysis Buffer. Chromatin stocks were sheared by sonication using a Qsonica sonicator on the high setting.
Chromatin Immunoprecipitation
The Chromatrap® ChIP-seq Protein A kit was used for ChIP according to the kit supplied protocol. Slurries were prepared for immunoprecipitation using 100 µl chromatin stock from each extraction with 2 µg H3K4me3 antibody or 2 µg rabbit IgG to determine nonspecific background binding.
An equivalent amount of chromatin for each IP was set aside as an input for use in downstream processes. Following reverse crosslinking and Proteinase K digestion, ChIP samples and inputs were purified using the Chromatrap® DNA purification columns supplied in the Chromatrap® ChIP-seq kit.
qPCR was carried out to determine the efficiency of precipitation of two gene loci using Chromatrap® primer sets for a positive gene target GAPDH and a negative gene target ß-globin.
At just 1000 cells, Chromatrap ChIP-seq kits proves sensitivity for low cell applications
Excellent signal was obtained for the occupancy of the histone mark H3K4me3 onto the positive target gene GAPDH (13%) when compared with non specific IgG and the negative gene target ß-globin.
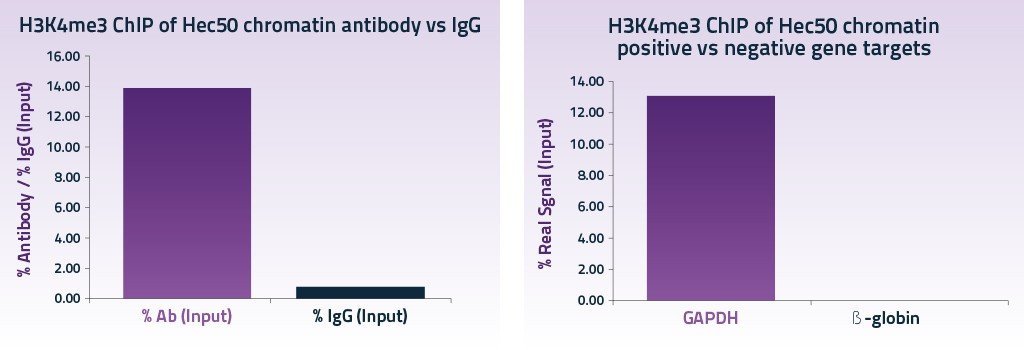
The low binding of H3K4me3 to the negative gene target ß-globin further emphasized the selectivity of the assay. The flexibility, specificity and sensitivity of the Chromatrap ChIP-seq Protein A kit allows robust and reproducible target enrichment from as little as 1000 cells per IP. Excellent signal strength is achieved and background noise is reproducibly low regardless of the number of cells used per IP.
Improve the quality of your research by equipping yourself with one of our Chromatrap® ChIP-seq kits - you can find the best one for you by consulting our step-by-step guide.

