
An informative list of everything you need to know about ChIP-Seq library preparation. The list provides a wealth of advice, from ensuring that the enzymes are stored at the appropriate temperature to collating analytical data using the Chromatrap® ChIP-seq Data Analysis Software.
1. Always use fresh reagents and consumables
High quality library preparation is key to a successful ChIP-seq experiment. It is important to prepare fresh reagents on the day of library preparation, especially those containing ethanol which can absorb water from the atmosphere. For gel excision always ensure a new clean scalpel is used. Library preparation kits contain enzymes with high rates of degradation at room temperature that affect their reaction efficiency. Ensure enzymes are stored at the appropriate temperatures throughout and return to cold storage immediately after use to preserve stability. To maximise sample recovery use low retention tubes which significantly reduce surface binding of DNA to the plasticware.
2. Use a fluorometer to quantify DNA
DNA must be accurately quantified before starting library preparation. Traditional spectrophotometers are not capable of accurately quantifying low concentrations of ChIP DNA. Fluorometric techniques that use intercalating dyes to quantify DNA are more accurate and have greater sensitivity than spectrophotometers. For the accurate quantification of DNA critical for ChIP and next generation sequencing (NGS) Chromatrap® recommend a fluorometric method.
3. Ensure high quality sheared chromatin between 100-300 bp
The success of any ChIP assay is highly dependent on the quality of the chromatin prepared. The chromatin must be sheared between 100-300 bp for ChIP-seq. Chromatrap® have found using a water bath sonicator with 30 second ON/OFF pulses for 15 minutes at a high power setting produces chromatin fragments of 100-300 bp. Ensure that the sample is kept at a stable 4ºC during the OFF phase. Shearing conditions will need to be optimised by the user prior to starting the immunoprecipitation. Further validation is recommended using a microfluidics platform such as the Agilent Bioanalyzer High Sensitivity DNA kit to ensure ChIP samples are of the expected size range and concentration.
4. Always use a ChIP-seq validated antibody
The quality of antibody is critical for generating premium data. Ensure that the antibody is ChIP-seq validated as antibodies verified for other applications may not work for NGS. A highly-specific antibody will increase the relative enrichment of the target, making it easier to detect binding events during data analysis. Many commercially available antibodies are listed as ChIP-seq grade and, wherever possible, should be used for your experiments. The Chromatrap® ChIP Seq kit provides a positive (H3K4me3) and a negative (IgG) antibody control in order to help validate your samples for sequencing. Chromatrap® also offers an antibody validation service, please contact Chromatrap® Customer Support for more information.
5. Use high sensitivity kits for library validation
At the beginning and end of library preparation your sample size distribution should be verified using an Agilent Bioanlyzer. Always use a high sensitivity DNA kit to ensure that DNA is accurately quantified and qualified. Remember that any adapters ligated during preparation will add weight to your sample and should be accounted for when performing size selection. For size selection Chromatrap® recommends the use of SYBR® Gold as the nucleic acid stain due to its sensitivity. Always leave an empty well between samples when loading the gel and remember to flank the samples with ladder to ensure that the gel has run evenly.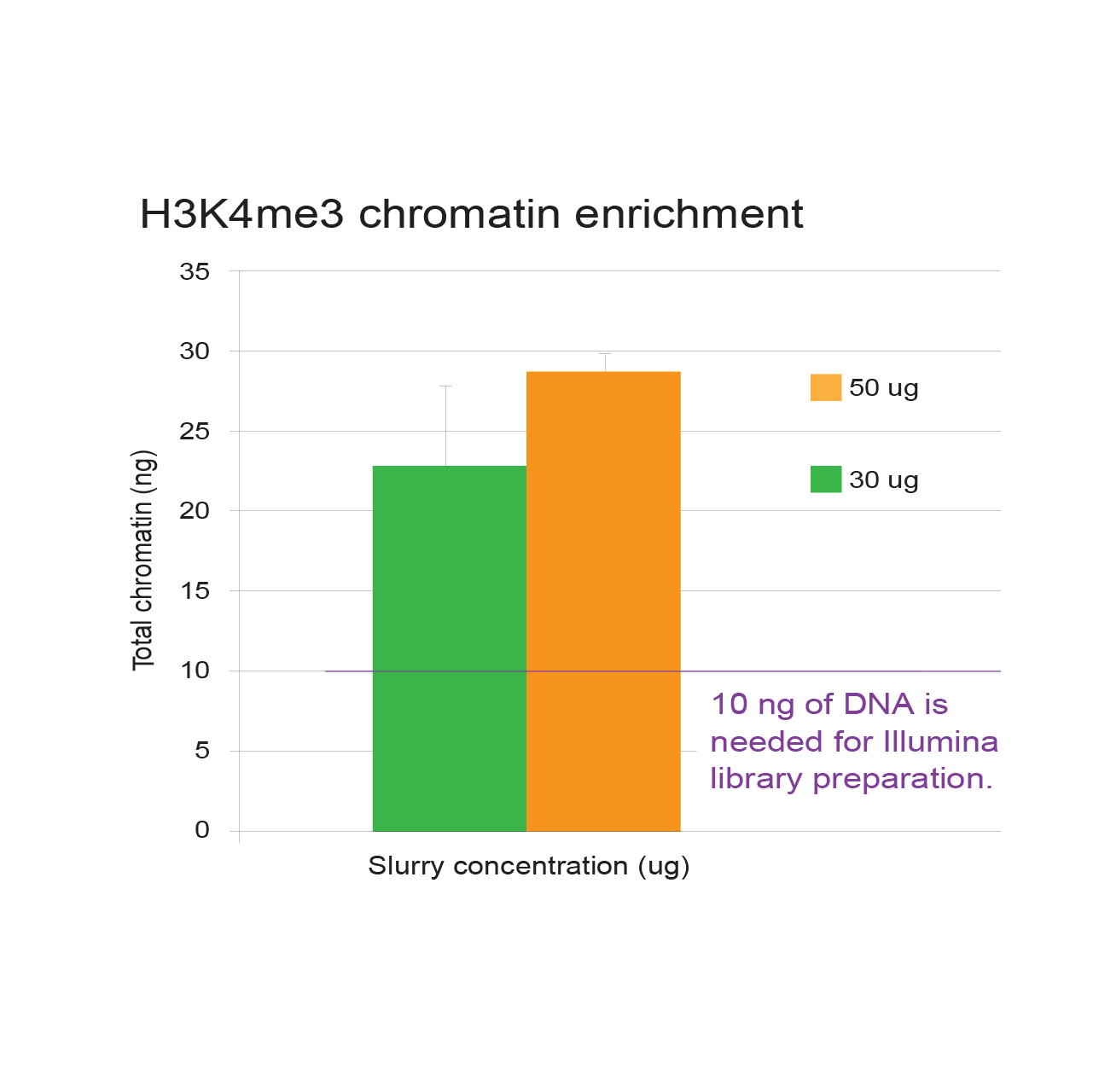
6. Quantity of DNA prior to library preparation
A typical ChIP experiment yields between 5-200 ng of DNA, requiring approximately 107 cells. The Illumina library preparation guide recommends using between 5-10 ng of ChIP DNA, however, we have found that increasing the starting quantity from 10 ng to 25 ng significantly reduces PCR duplication bias. Chromatrap® therefore recommend library synthesis using 25 ng of IP DNA.
7. Always prepare controls
Peaks identified during sequence analysis must be compared to the same region in a matched control sample in order to verify their significance. For example, a random region of repetitive sequences may appear enriched due to the number of copies of the region, creating a false-positive result. There are three commonly used controls: input DNA (DNA that has not been immunoprecipitated); mock IP (DNA treated the same but without antibody during the IP); and non-specific IP (IP with an antibody targeting a protein not known to be involved in DNA binding such as IgG). There is no consensus as to which control is most appropriate to use, however, Chromatrap® recommends using input DNA as this accounts for bias related to shearing and is commonly used in peer reviewed journals.
8. Always validate by qPCR before sequencing
Before beginning library synthesis, we recommend analysing the immunoprecipitated DNA by qPCR using at least one gene-specific positive and one negative control target of your choice. Chromatrap® provides a positive control primer set for GAPDH which has been validated for use with the positive control antibody H3K4me3, to validate your results.
9. Magnetic bead preparation
All library preparation kits use magnetic beads which should always be vortexed thoroughly immediately before use and equilibrated to room temperature. Do not over dry the beads as this can make re-suspension difficult and hamper appropriate elution. When the beads start to show “cracks” within the pellet this is a good time to resuspend.
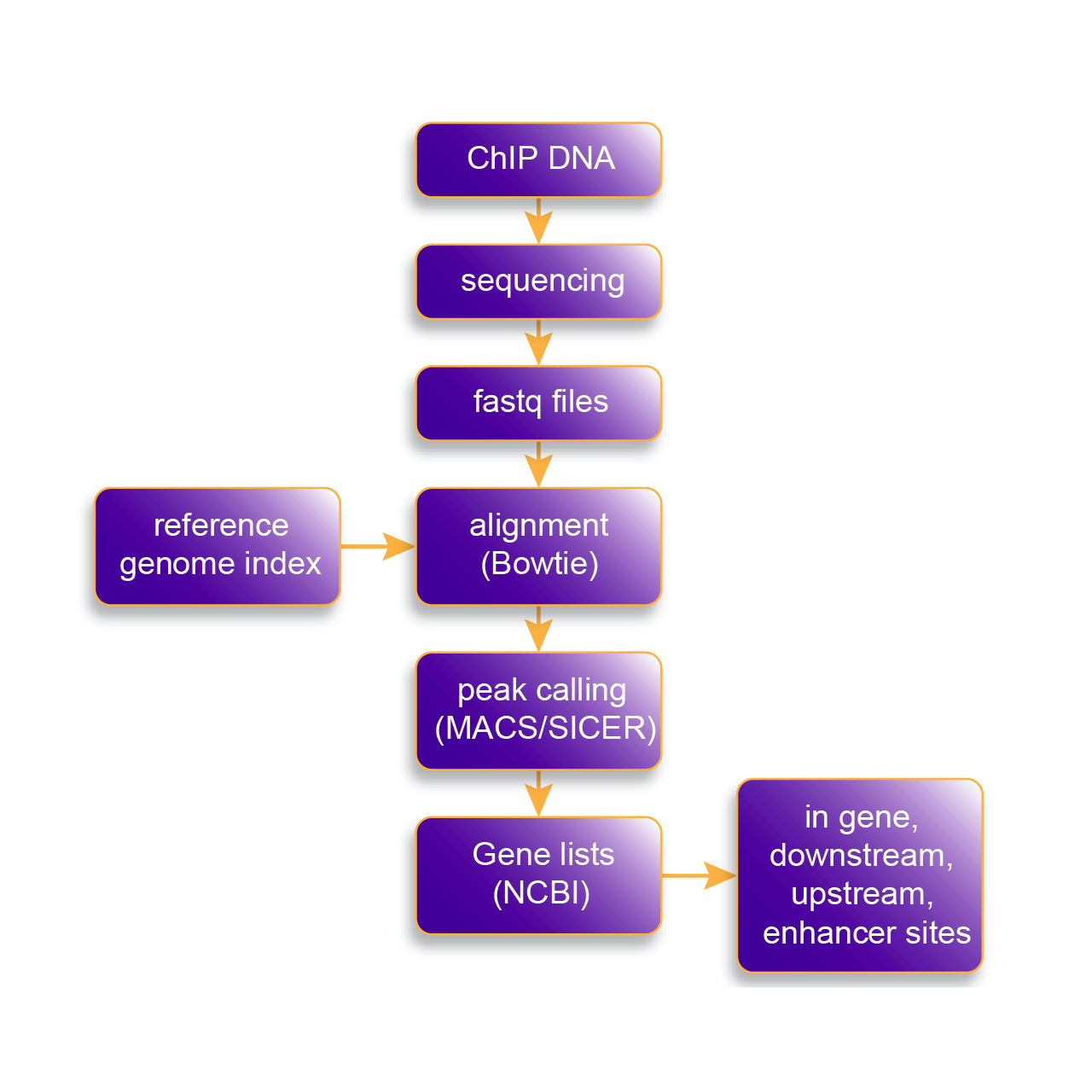
10. Sound analysis pipeline
Reads must be mapped to a reference genome before peak-calling software is used to identify regions of enrichment. An initial impression of ChIP-seq quality can be obtained by the inspection of mapped sequence reads within a known binding location. More detailed analysis requires the use of dedicated software algorithms and The Chromatrap® ChIP-seq Data Analysis Software allows read alignment and peak calling for narrow and broad peak marks such as transcription factors and histone modifications. Users can also compare gene lists to identify specific targets that are uniquely or commonly enriched among their dataset.
Make sure your ChIP-Seq library preparation is at the highest standard with one of our revolutionary Chromatrap® ChIP-seq kits or get in touch with our team for any queries.
